Ⅰ. Introduction
Precise Drug Target Identification is a first step for improving efficacy,
tracing and avoiding side-effects as well as understanding the mode-of-actions of drug candidates.
However, until now, effective systematic approaches for the precise drug target identification at genome levels
have not been commercially available. Using S. pombe genome-wide heterozygous deletion mutant library,
Bioneer has developed a high-throughput genome-wide drug target screening service system (GPScreen™) and
made it commercially available for your drug discovery needs. GPScreen™ screening service covers a broad spectrum
of drug candidates in whole disease areas from cancers & metabolic diseases to neglected & rare diseases and, therefore,
will provide the total solutions for an efficient drug discovery through providing clear-cut answers to problems such
as drug-target(s) and toxicity as well as mode-of-actions of drug candidates.
“Bioneer’s GPScreen™ using S. pombe genome-wide heterozygous deletion mutant library
has enabled genome-wide screening for drug target identification”
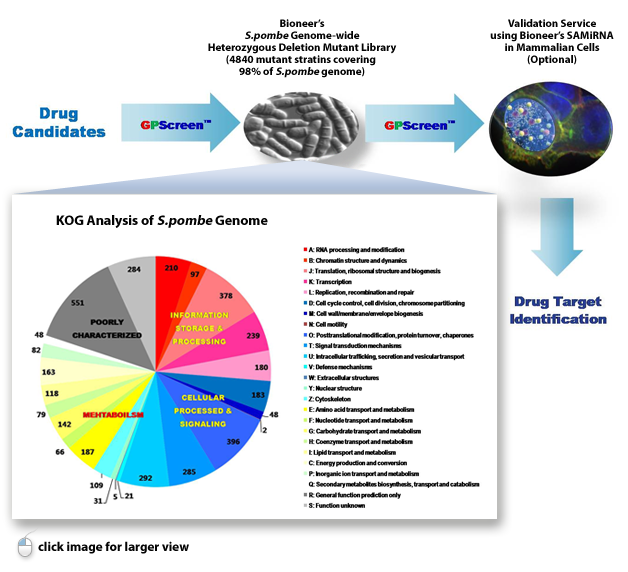
GPScreen™ is based on Bioneer’s unique S. pombe genome-wide deletion mutant library
(http://pombe.bioneer.com/), which was developed by Bioneer and the Korea Research Institute of Bioscience & Biotechnology
(KRIBB) in collaboration with Dr. Paul Nurse of Cancer Research Center UK (Nat. Biotech, 28, 617–623 (2010)).
It can be used for genetic and chemical screening such as drug target identification,
gene expression profiling and synthetic lethal profiling.
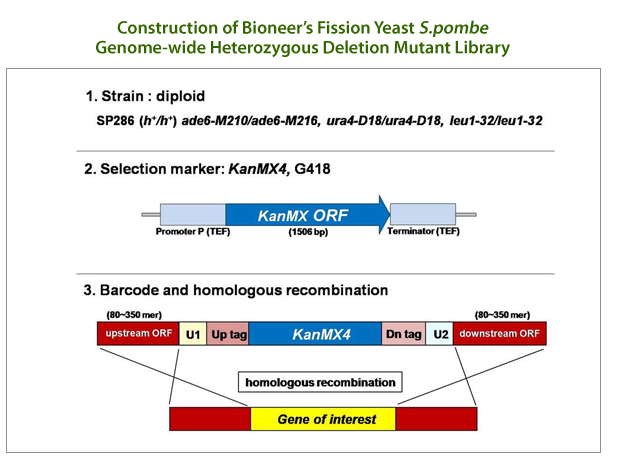
This mutant library comprised of a total of 4,840 heterozygous diploid deletion mutants representing 98.5%
genome coverage and one copy of a specific gene in each mutant was deleted by homologous recombination.
GPScreen™ exclusively offers S. pombe deletion mutant library as a powerful tool for large-scale genetic functional
analysis, identification and verification research of drug targets and for integrated systems research of cell function.
Ⅱ. Principle of GPScreen™
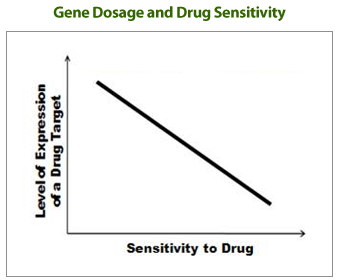
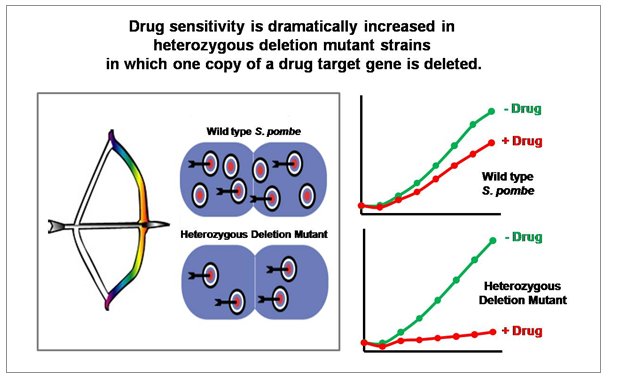
“Drug-induced Haploinsufficiency (DIH)” is a situation in which a strain with a heterozygous gene deletion
(producing target proteins to about half of normal levels), result in cells sensitive to a specific drug.
DIH occurs when a drug or drug candidate acts on a mutant strain in which the level of target gene protein is lowered from normal
to half level, and is considered to be a valuable tool for drug target identification. Previously,
a number of reports have provided identifications of drug targets using DIH in budding yeast
S. cereviase (Baetz K et al., 2004; Lum et al., 2004).
However, fission yeast S. pombe is considered a better model of mammalian cells since its cell cycle regulation is closer to that of mammalian cells than that of S. cerevisiae.
“GPScreen™ utilizes DIH in the fission yeast S. pombe genome-wide heterozygous deletion mutant library to identify the drug target(s).
References
• Baetz K et al. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. PNAS. 2004
[More information]
• Lum PY et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes.
Cell. 2004
[More information]
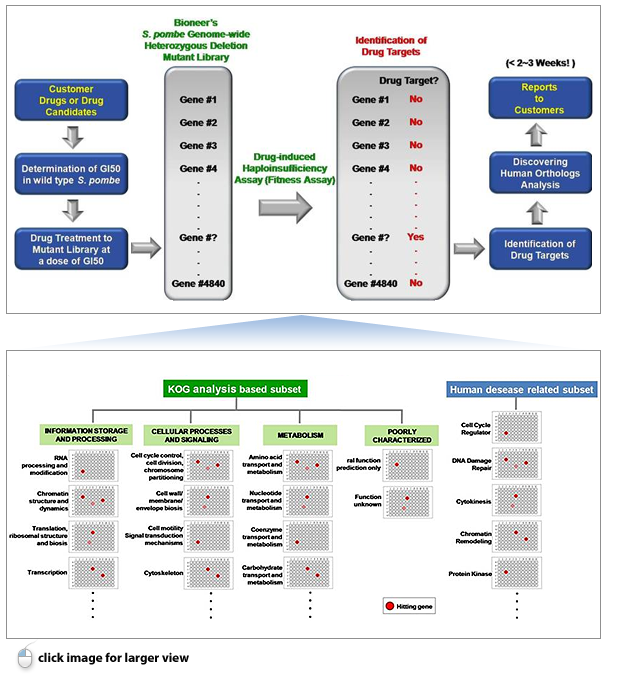
Ⅲ. Performance of GPScreen™
GPScreen™ is a best-in class genome-wide drug target identification system that is exclusively offered by Bioneer Inc. It allows for the clear-cut identification of molecular targets of drugs/ drug candidates (A representative is below).
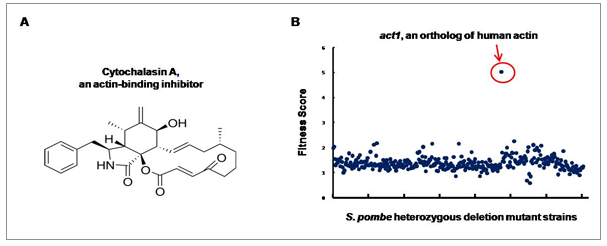 Figure 1. Drug Target Identification using GPScreen™.
Figure 1. Drug Target Identification using GPScreen™.
In the presence of cytochalasin A (Fig. 1A), a heterozygous deletion mutant of act1 shows decreased growth as a result of a decrease in “functional” Act1 protein. act1 was the only gene in the genome-wide screen to show this effect, demonstrating that act1 is a target of cytochalasin A (Fig. 1B).

Ⅳ. Flow chart of GPScreen™ Service